
Rhizopus

| Rhizopus | |
|---|---|
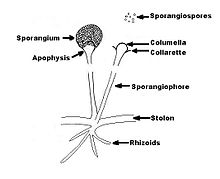
| |
| Schematic diagram of Rhizopus spp. | |
| Scientific classification | |
| Domain: | Eukaryota |
| Kingdom: | Fungi |
| Division: | Mucoromycota |
| Class: | Mucoromycetes |
| Order: | Mucorales |
| Family: | Mucoraceae |
| Genus: | Rhizopus Ehrenb. (1820) |
| Type species | |
| Rhizopus nigricans Ehrenb. (1820)
| |
| Synonyms[1] | |
Rhizopus is a genus of common saprophytic fungi on plants and specialized parasites on animals. They are found in a wide variety of organic substances, including "mature fruits and vegetables",[2] jellies, syrups, leather, bread, peanuts, and tobacco. They are multicellular. Some Rhizopus species are opportunistic human pathogens that often cause fatal disease called mucormycosis. This widespread genus includes at least eight species.[3][4]

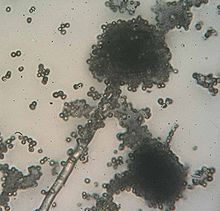
Rhizopus species grow as filamentous, branching hyphae that generally lack cross-walls (i.e., they are coenocytic). They reproduce by forming asexual and sexual spores. In asexual reproduction, sporangiospores are produced inside a spherical structure, the sporangium. Sporangia are supported by a large apophysate columella atop a long stalk, the sporangiophore. Sporangiophores arise among distinctive, root-like rhizoids. In sexual reproduction, a dark zygospore is produced at the point where two compatible mycelia fuse. Upon germination, a zygospore produces colonies that are genetically different from either parent.

- Rhizopus oligosporus is used to make tempeh, a fermented food derived from soybeans.
- Rhizopus oryzae is used in the production of alcoholic beverages in parts of Asia and Africa.
- Rhizopus stolonifer (black bread mold) causes fruit rot on strawberry, tomato, and Sweet potato and is used in commercial production of fumaric acid and cortisone.
Various species, including R. stolonifer, may cause soft rot in sweet potatoes and Narcissus.

Rhizopus helps in nutrient development since this species is grown in soil it ferments the fruits and vegetable in the soil inhibiting the growth and develops certain pathogens that inhibits the growth of toxigenic fungus.[5] In addition to that, there is even a type of Rhizopus (Rhizopus microsporus-fermented soybean tempe) that has proven to reduce colon carcinogenesis in rats by elevating factors of mucins, immunoglobulin A, and organic acids and give protection to piglets from Escherichia coli-infection by inhibiting adhesion to the intestinal membranes. [6]

Phylogeny
The mating analysis has also been found which was comparative that this species structure is flexible in comparison with other species in the same genus. The topology length of the species genome is found to be three times bigger with the species.[7]

Species
- Rhizopus americanus
- Rhizopus arrhizus
- Rhizopus azygosporus
- Rhizopus caespitosus
- Rhizopus circinans
- Rhizopus delemari
- Rhizopus homothallicus
- Rhizopus lyococcus
- Rhizopus microsporus
- Rhizopus niveus
- Rhizopus oligosporus
- Rhizopus oryzae
- Rhizopus reflexus
- Rhizopus rhizopodiformis
- Rhizopus schipperae
- Rhizopus sexualis
- Rhizopus stolonifer (black bread mold)
See also
References
- ^ "Rhizopus Ehrenb. 1820". MycoBank. International Mycological Association. Retrieved 2011-02-05.
- ^ Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008). Dictionary of the Fungi (10th ed.). Wallingford, UK: CABI. p. 599. ISBN 978-0-85199-826-8.
- ^ Zheng RY, Chen GQ, Huang H, Liu XY (2007). "A monograph of Rhizopus". Sydowia. 59 (2): 273–372.
- ^ Abe A, Asano K, Sone T (2010). "A Molecular Phylogeny-Based Taxonomy of the Genus Rhizopus". Bioscience, Biotechnology, and Biochemistry. 74 (7): 1325–1331. doi:10.1271/bbb.90718. PMID 20622457. S2CID 13369408.
- ^ Dwi Endrawati, Dwi Endrawati. "Several Functions of Rhizopus sp on Increasing Nutritional Value of Feed Ingredient". June 2017.
- ^ Yang, Yongshou (2018). "The effects of tempe fermented with Rhizopus microsporus, Rhizopus oryzae, or Rhizopus stolonifer on the colonic luminal environment in rats". Journal of Functional Foods. 49: 162–167. doi:10.1016/j.jff.2018.08.017. S2CID 91791814.
- ^ Gryganskyi, Andrii P; Golan, Jacob; Dolatabadi, Somayeh; Mondo, Stephen; Robb, Sofia; Idnurm, Alexander; Muszewska, Anna; Steczkiewicz, Kamil; Masonjones, Sawyer; Liao, Hui-Ling; Gajdeczka, Michael T; Anike, Felicia; Vuek, Antonina; Anishchenko, Iryna M; Voigt, Kerstin; de Hoog, G Sybren; Smith, Matthew E; Heitman, Joseph; Vilgalys, Rytas; Stajich, Jason E (1 June 2018). "Phylogenetic and Phylogenomic Definition of Rhizopus Species". G3: Genes, Genomes, Genetics. 8 (6): 2007–2018. doi:10.1534/g3.118.200235. PMC 5982828. PMID 29674435.
External links
- Rhizopus at Zygomycetes.org
- Photos of Rhizopus spp. used for tempeh-making[permanent dead link] at www.tempeh.idv.tw

See what we do next...
OR
By submitting your email or phone number, you're giving mschf permission to send you email and/or recurring marketing texts. Data rates may apply. Text stop to cancel, help for help.
Success: You're subscribed now !
