
Zoopagomycotina

| Zoopagomycotina | |
|---|---|

| |
| Fertile heads of Piptocephalis | |
| Scientific classification | |
| Domain: | Eukaryota |
| Kingdom: | Fungi |
| Division: | Zoopagomycota |
| Subdivision: | Zoopagomycotina Benny, 2007 |
| Class: | Zoopagomycetes Doweld, 2014 |
| Order and families | |
The Zoopagomycotina are a subdivision (incertae sedis) of the fungal division Zygomycota sensu lato.[1] It contains 5 families and 20 genera.[2] Relationships among and within subphyla of Zygomycota are poorly understood, and their monophyly remains in question, so they are sometimes referred to by the informal name zygomycetes.

Zoopagomycotina are microscopic and are typically obligate parasites of other zygomycete fungi and of microscopic soil animals such as nematodes, rotifers and amoebae.[3] Some species are endoparasites that live mostly within the bodies of their hosts and only exit the host when they are producing spores. Other species are ectoparasites (e.g. Syncephalis, Piptocephalis)[2] that live outside of the host body but produce specialized organs called haustoria that penetrate inside of the host body to capture host nutrients. Similar haustoria are found in biotrophic plant, animal and fungal pathogens in several other major fungal lineages.

Like most other zygomycete fungi, the Zoopagomycotina have cell walls containing chitin and have coenocytic (nonseptate) hyphae. Their vegetative body consists of a simple, branched or unbranched thallus. Asexual reproduction is by arthrospores (in Helicocephalum), chlamydospores, uni- or multi-spored sporganiola; sporangiospores of multi-spored formed in simple or branched chains (merosporangia), usually from a vesicle or stalk. Many produce haustoria. Where observed, the sexual spores (zygospores) are globose and unornamented. The hyphae used during sexual outcrossing is similar to vegetative hyphae or in some cases may be slightly enlarged.[1]

Etymology
The word Zoopagomycotina comes from the Greek roots zoo meaning "animal" and pag meaning "rock" or "ice/frost".[4]

Evolutionary relationships

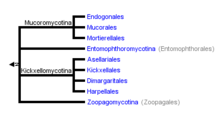
Although great strides have been made in resolving the evolutionary relationships among many lineages of fungi,[5] it has been challenging to resolve relationships within and among zygomycetes. For example, the uncertain grouping of Zoophagus insidians with the Kickxellomycotina.[1]

Resolving a well-supported monophyly of the Zoopagomycotina has been particularly challenging for several main reasons:

- most species of Zoopagomycotina are microscopic and challenging to observe
- most species of Zoopagomycotina cannot be grown separately from their host organisms in axenic culture, so obtaining pure DNA for molecular studies is challenging
- based on ribosomal DNA sequences, species of Zoopagomycotina may have undergone accelerated evolution, so grouping may be skewed by long-branch attraction (LBA) and a high frequency of parallel evolutionary changes[6]
Families and their respective genera
- Class Zoopagomycetes Doweld 2014
- Order Zoopagales Bessey 1950 ex. Benjamin 1979 [Zoophagales Doweld 2014]
- Massartia De Wildeman 1897 non Conrad 1926
- Family Basidiolaceae Doweld 2013
- Basidiolum Cienk. 1861
- Family Cochlonemataceae Duddington 1974
- Amoebophilus (6 spp.) ectoparasite of amoebae
- Aenigmatomyces Castan˜eda & Kendrick 1993
- Aplectosoma (1 sp.) parasite of an amoeba
- Bdellospora (1 sp.) ectoparasite of an amoeba
- Cochlonema (19 spp.) endoparasites of amoebae and rhizopods
- Endocochlus (4 spp.) endoparasites of amoebae
- Euryancale (5 spp. ) endoparasites of nematodes
- Family Helicocephalidaceae Boedijn 1959
- Brachymyces (1 sp.) parasitic on bdelloid rotifers
- Helicocephalum (5 spp.) parasites of small animals, especially nematodes and nematode eggs
- Rhopalomyces (8 spp.) parasites of small animals, especially nematodes and nematode eggs
- Verrucocephalum Degawa 2013
- Family Piptocephalidaceae Schröter 1886
- Kuzuhaea (1 sp.) haustorial parasite of fungi (mostly of Mucorales spp.)
- Piptocephalis (25 spp.) haustorial parasite of fungi (mostly of Mucorales spp.)
- Syncephalis (61 spp.) haustorial parasite of fungi (mostly of Mortierellales and Mucorales spp.)
- Family Sigmoideomycetaceae Benny, Benjamin & Kirk 1992
- Reticulocephalis (2 spp.) putative haustorial parasite of fungi
- Sigmoideomyces (2 spp.) putative haustorial parasite of fungi
- Sphondylocephalum Stalpers 1974
- Thamnocephalis (3 spp.) haustorial parasite of fungi
- Family Zoopagaceae Drechsler 1938
- Acaulopage (27 spp.) haustorial parasites of amoeba
- Cystopage (7 spp.) haustorial parasites of amoebae and nematodes
- Lecophagus Dick 1990
- Stylopage (18 spp.) predaceous on amoebae and nematodes
- Tentaculophagus Doweld 2014
- Zoopage (11 spp.) haustorial parasite of amoebae and testaceous rhizopods
- Zoophagus (5 spp.) ectoparasites of loricate rotifers and nematodes[2]
- Order Zoopagales Bessey 1950 ex. Benjamin 1979 [Zoophagales Doweld 2014]
References
- ^ a b c Hibbett DS, Binder M, Bischoff JF, et al. (May 2007). "A higher-level phylogenetic classification of the Fungi". Mycol. Res. 111 (Pt 5): 509–47. CiteSeerX 10.1.1.626.9582. doi:10.1016/j.mycres.2007.03.004. PMID 17572334.
- ^ a b c url=http://zygomycetes.org/index.php?id=8
- ^ url=http://www.tolweb.org/Zygomycota
- ^ url=http://www.oed.com
- ^ James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, et al. (2006). "Reconstructing the early evolution of Fungi using a six-gene phylogeny". Nature. 443 (Pt 7113): 818–822. Bibcode:2006Natur.443..818J. doi:10.1038/nature05110. PMID 17051209. S2CID 4302864.
- ^ White MM, et al. (2006). "Phylogeny of the Zygomycota based on nuclear ribosomal sequence data". Mycologia. 98 (Pt 6): 872–884. doi:10.3852/mycologia.98.6.872. PMID 17486964.
External links
See what we do next...
OR
By submitting your email or phone number, you're giving mschf permission to send you email and/or recurring marketing texts. Data rates may apply. Text stop to cancel, help for help.
Success: You're subscribed now !
